---
title: "Covid-19 Trends in the Netherlands"
description: "Based on data published by RIVM, in this post I looked at how Covid-19 cases spread throughout the Netherlands."
author: "Muhammad Chenariyan Nakhaee"
date: "2021-05-22"
categories: [R, data visualization, Covid-19]
image: featured.JPG
format:
html:
code-fold: true
code-tools: true
toc: true
---
```{r message=FALSE, warning=FALSE, include=FALSE}
knitr::opts_chunk$set(
echo = TRUE,
message = FALSE,
warning = FALSE,
eval = FALSE
)
# Font configuration - uncomment if fonts are installed
# windowsFonts('Lobster' = 'Lobster')
# windowsFonts('Poppins Light' = 'Poppins Light')
```
Two weeks ago, I made [a visualization](https://www.linkedin.com/posts/muhammadcnakhaee_covid19-datavisualization-ggplot-activity-6726775611223764992-c_dk) that shows how Covid-19 cases spread in the Netherlands from the beginning of March and how grim the situation looked. However, someone pointed out the fact that the number of tests has increased significantly. It means that my plot may exaggerate the Covid-19 situation in the Netherlands. Unfortunately, I could not find testing data for each Dutch municipality. Instead, I decided to use hospitalization admissions and deceased cases to see if we can indeed see a massive spread in the second wave of Covid-19 cases in the Netherlands.
```{r,eval=FALSE}
library(tidyverse)
library(CoronaWatchNL)
library(sf)
library(gganimate)
library(santoku)
library(lubridate)
library(foreign)
theme_set(theme_void())
theme_update(
#plot.background = element_rect(fill = '#FDF6E3',color = '#FDF6E3'),
text = element_text(family = 'Poppins Light'),
plot.subtitle = element_text(
family = 'Poppins Light',
size = 10,
margin = margin(b = 10)
),
plot.title = element_text(
family = 'Poppins Light',
size = 12,
margin = margin(t = 10, b = 10)
)
)
```
I created an R package called [CoronaWatchNL](https://github.com/mcnakhaee/CoronaWatchNL) that allows you to access a wide range of Covid-19 datasets. I'll use this package in this post to get Covid-19 cases, hospital admissions, and deaths for Dutch municipalities.
::: {.callout-note}
This post uses R packages that need to be installed: `tidyverse`, `CoronaWatchNL`, `sf`, `gganimate`, `santoku`, `lubridate`, `foreign`. The code is shown for educational purposes but not executed in this rendered version.
:::
```{r warning=FALSE, message=FALSE,eval=FALSE}
municipalBoundaries <- st_read(
"https://geodata.nationaalgeoregister.nl/cbsgebiedsindelingen/wfs?request=GetFeature&service=WFS&version=2.0.0&typeName=cbs_gemeente_2020_gegeneraliseerd&outputFormat=json"
)
daily_cases_per_municpality <- get_daily_cases_per_municipality()
populatuon_per_region <- get_population_per_region()
daily_cases_per_municpality <- daily_cases_per_municpality %>%
inner_join(populatuon_per_region, by = c('Municipality_name' = 'Regions')) %>%
mutate(
Date_of_publication = as_date(Date_of_publication),
avg_daily_total_cases = 100000 * as.numeric(Total_reported) / as.numeric(`Bevolking op 1 januari (aantal)`),
avg_daily_hospital_admissions = 100000 * as.numeric(Hospital_admission) / as.numeric(`Bevolking op 1 januari (aantal)`),
avg_daily_deceased = 100000 * as.numeric(Deceased) / as.numeric(`Bevolking op 1 januari (aantal)`)
)
```
I compute the weekly average number of Covid-19 cases, hospitalizations, and deaths per 100,000 inhabitants in each municipality in the Netherlands. The following piece of code shows how I did this using R.
```{r fig.height=10,fig.width=10,eval=FALSE}
weekly_cases <- daily_cases_per_municpality %>%
mutate(week = round_date(Date_of_publication , unit = 'week'))
weekly_cases_per_municpality <- weekly_cases %>%
group_by(Municipality_name, week) %>%
summarise(
avg_weekly_total_cases = mean(avg_daily_total_cases),
avg_weekly_hospital_admissions = mean(avg_daily_hospital_admissions),
avg_weekly_deceased = mean(avg_daily_deceased)) %>%
ungroup() %>%
mutate(
d_avg_weekly_total_cases = chop(avg_weekly_total_cases, c(0, 0, 0.5, 1, 5, 12, 20, 35, 55, 80, 100)),
d_avg_hospital_admissions = chop(
avg_weekly_hospital_admissions,
c(0, 0, 0.5, 1, 2, 3, 5, 7, 9, 10, 15)),
d_avg_weekly_deceased = chop(avg_weekly_deceased, c(0, 0, 0.1, 0.5, 1, 1.5, 2, 2.5, 3, 5)))
data_weekly <- municipalBoundaries %>%
right_join(weekly_cases_per_municpality,
by = c(statnaam = "Municipality_name"))
```
I will create an animation that shows how Covid-19 cases spread in the Netherlands and which municipalities were and are hit hardest by the pandemic.
```{r eval=FALSE}
make_animation <- function(data, var_name, pal, title) {
var_name <- rlang::enquo(var_name)
data %>%
#filter(week > '2020-10-01') %>%
ggplot() +
geom_sf(aes(fill = !!var_name), color = 'gray95') +
scale_fill_manual(values = pal) +
coord_sf(datum = NA) +
labs(
title = title,
subtitle = 'Date: {current_frame}',
fill = 'Counts per 100,000',
caption = 'Source: RIVM'
) +
transition_manual(week, cumulative = T) +
ease_aes("sine") +
enter_fade(alpha = 0.5) +
exit_fade(alpha = 0.5)
}
```
## Covid-19 Cases
The first animation shows the number of infections in each municipality, from the start of the pandemic in February until recently. As you can see, the second wave, which began in late September, looks really terrifying. Note that there are some municipalities for which no data is available.
```{r eval=FALSE}
pal_cases <- c(
'gray95',
'#fee440',
'#FFBA08',
'#FAA307',
'#F48C06',
'#E85D04',
'#DC2F02',
'#D00000',
'#9D0208',
'#6A040F',
'#370617',
'#03071e'
)
make_animation(data_weekly,d_avg_weekly_total_cases,pal_cases,'The Average Weekly Number of Covid-19 Cases\nper 100,000 Inhabitants in the Netherlands')
```
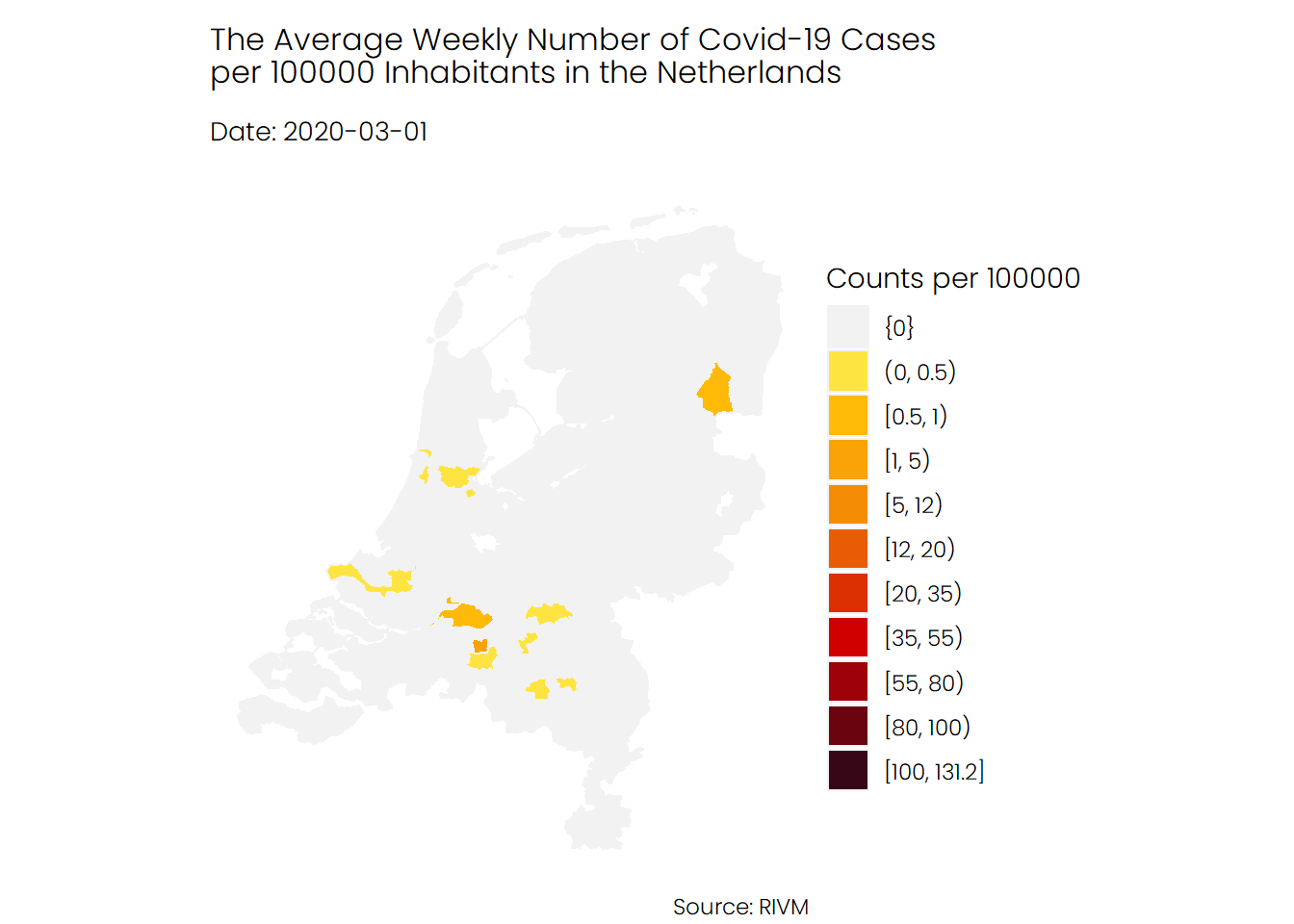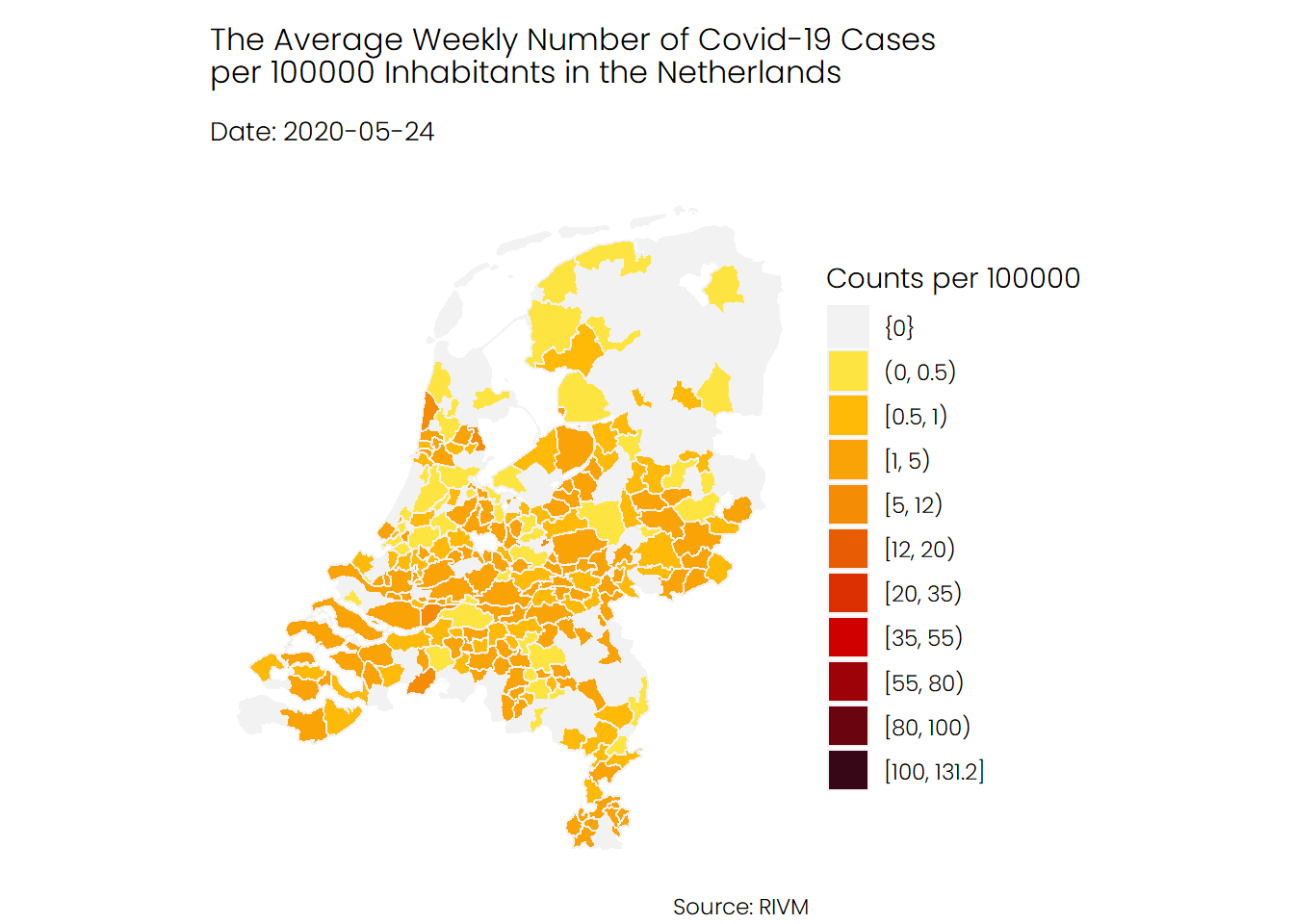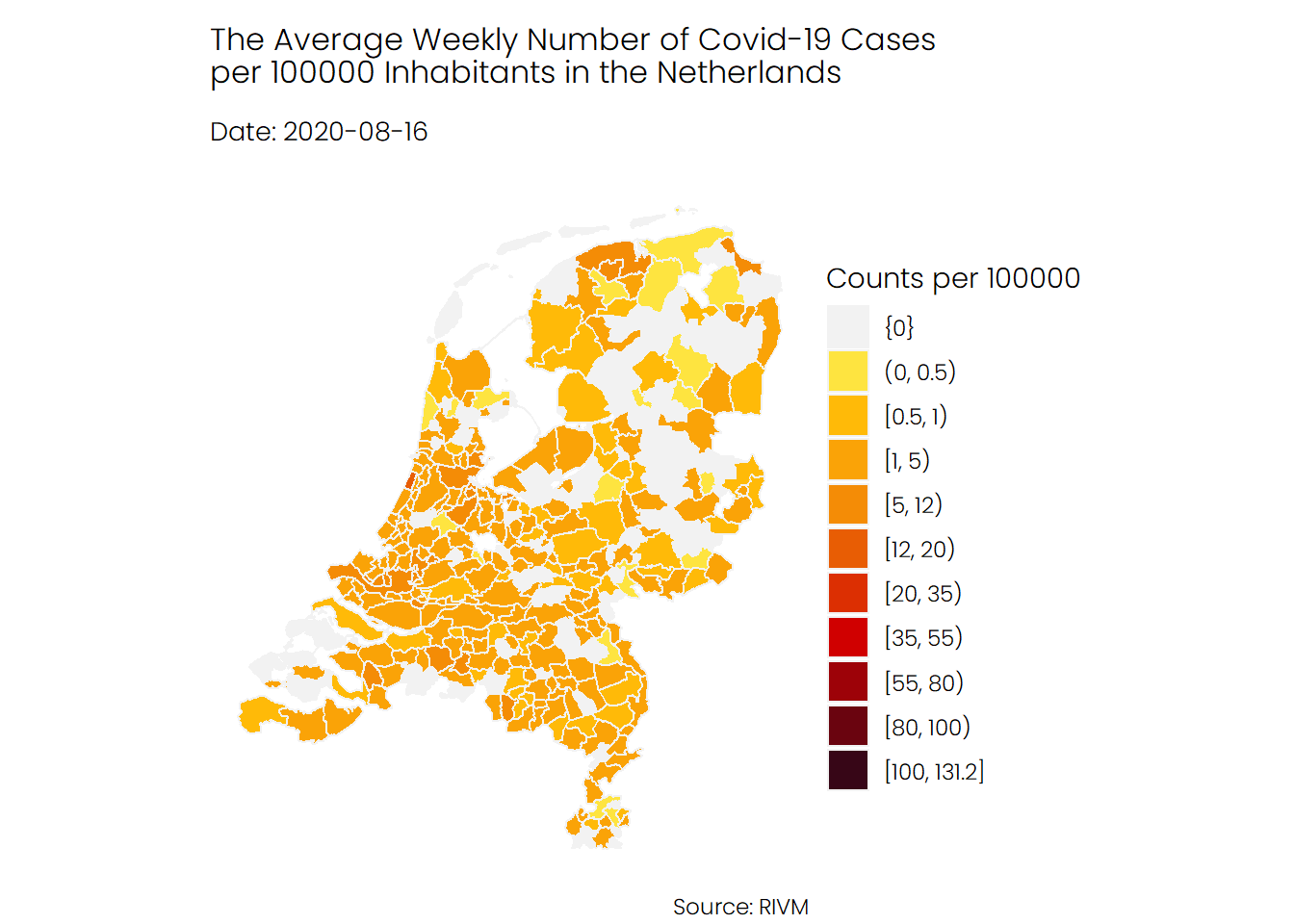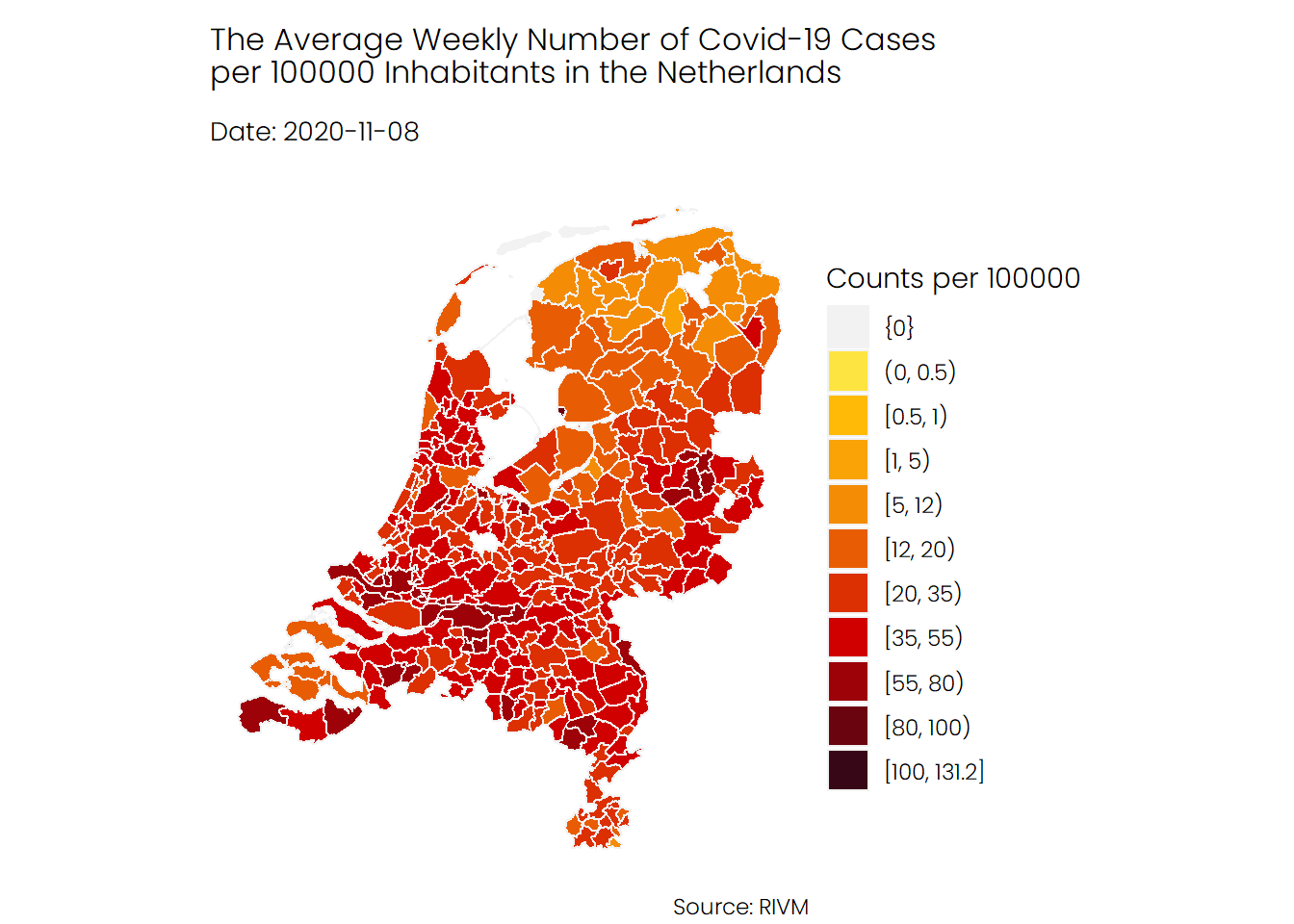
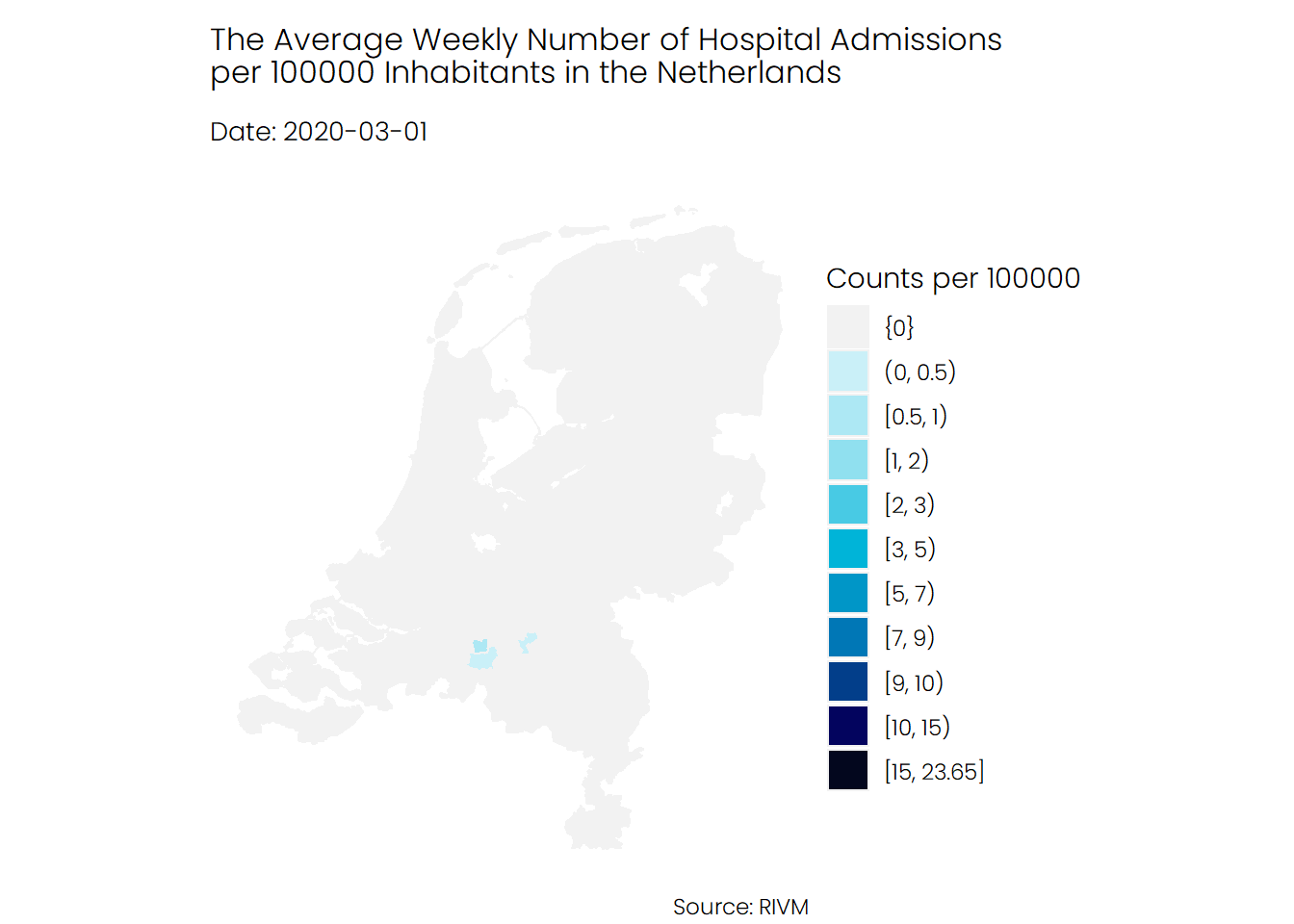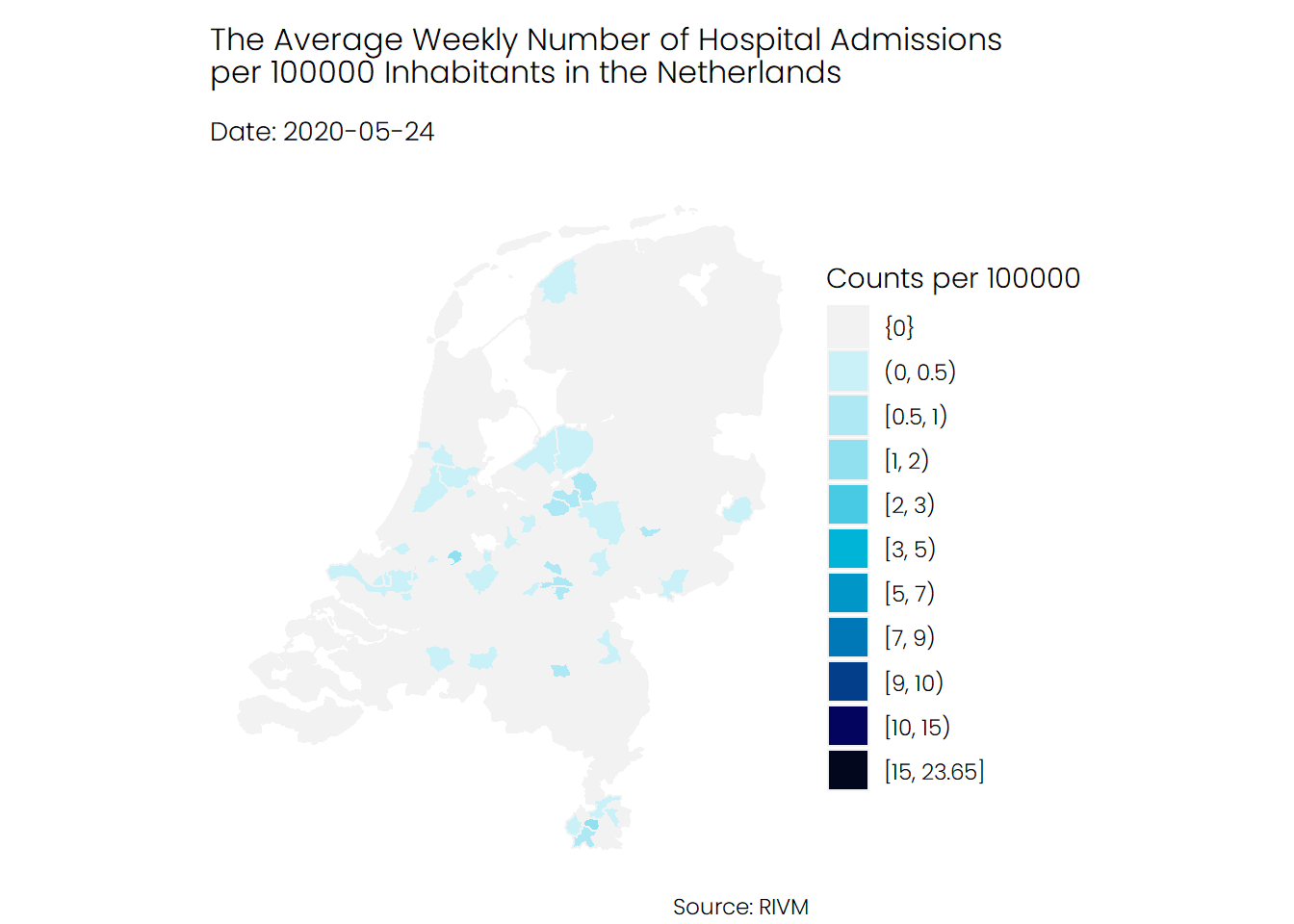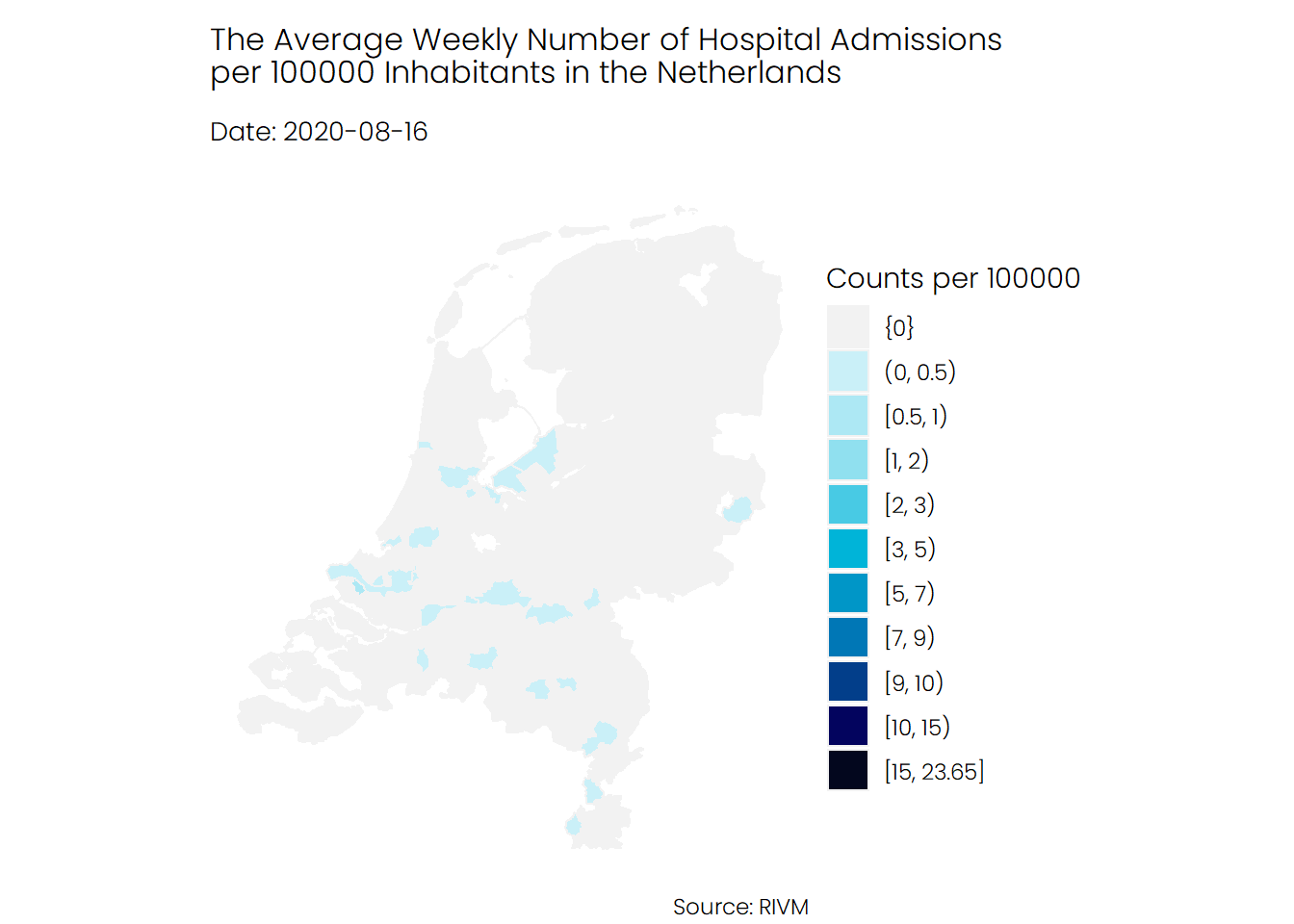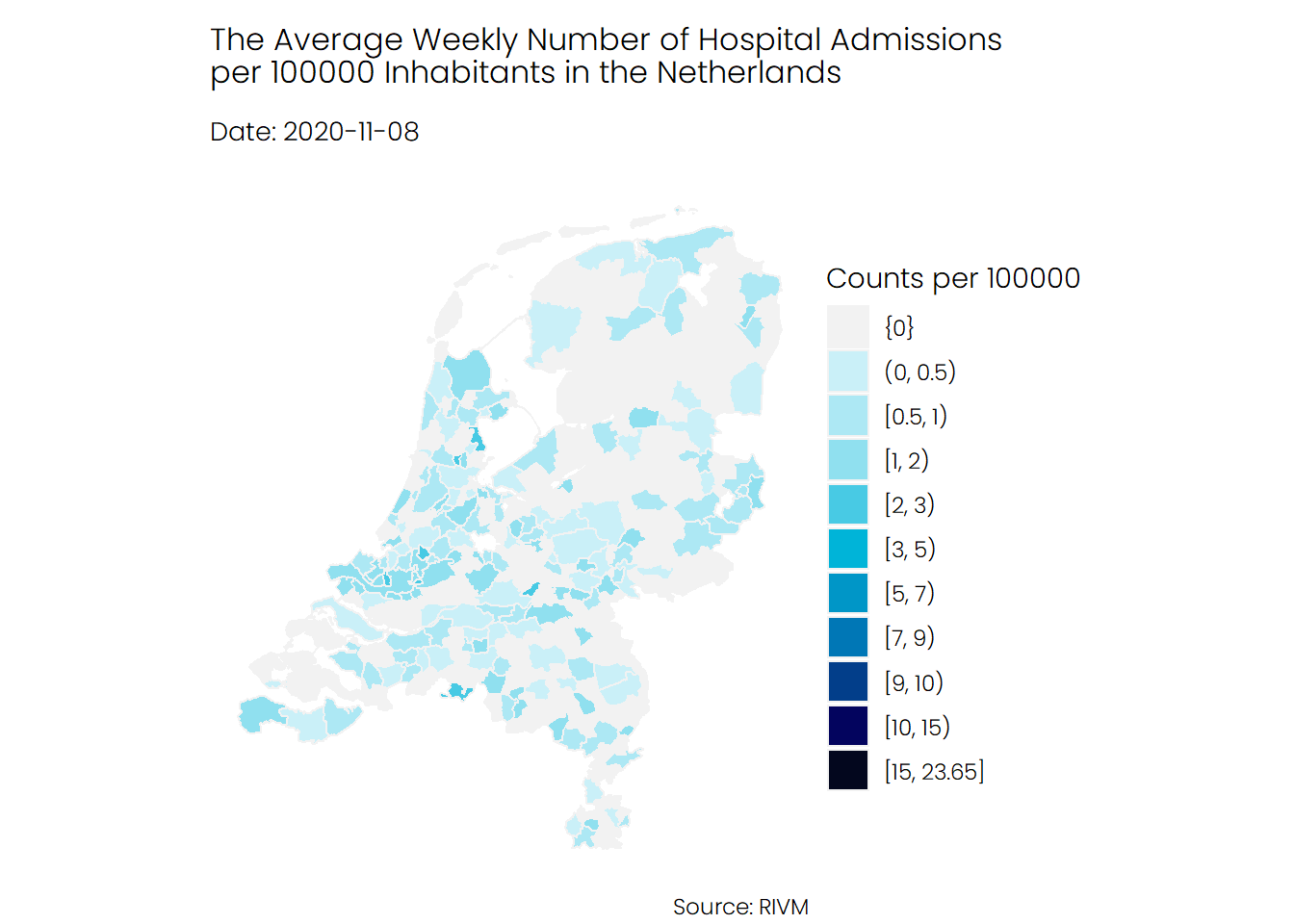
## Hospital Admissions
If we look at the number of hospital admissions, we see a different story. It seems that the number of hospitalizations was higher during the first wave of Covid-19 compared to the second wave, and mostly the southern parts of the Netherlands were hit harder than the rest of the Netherlands.
```{r eval=FALSE}
pal_patients <- c(
'gray95',
'#caf0f8',
'#ade8f4',
'#90e0ef',
'#48cae4',
'#00b4d8',
'#0096c7',
'#0077b6',
'#023e8a',
'#03045e',
'#03071e'
)
make_animation(data_weekly,d_avg_hospital_admissions,pal_patients,'The Average Weekly Number of Hospital Admissions\nper 100,000 Inhabitants in the Netherlands')
```
## Deaths
The trend for deceased patients looks similar to that of hospital admissions. The average number of deaths during the first wave of coronavirus was higher than the average number of deaths during the second wave.
```{r eval=FALSE}
pal_deceased <- c(
'gray95',
'#fdc5f5',
'#e0aaff',
'#c77dff',
'#9d4edd',
'#7b2cbf',
'#5a189a',
'#3c096c',
'#240046',
'#10002b'
)
make_animation(data_weekly,d_avg_weekly_deceased,pal_deceased,'The Average Weekly Number of Deceased Patients\nper 100,000 Inhabitants in the Netherlands')
```
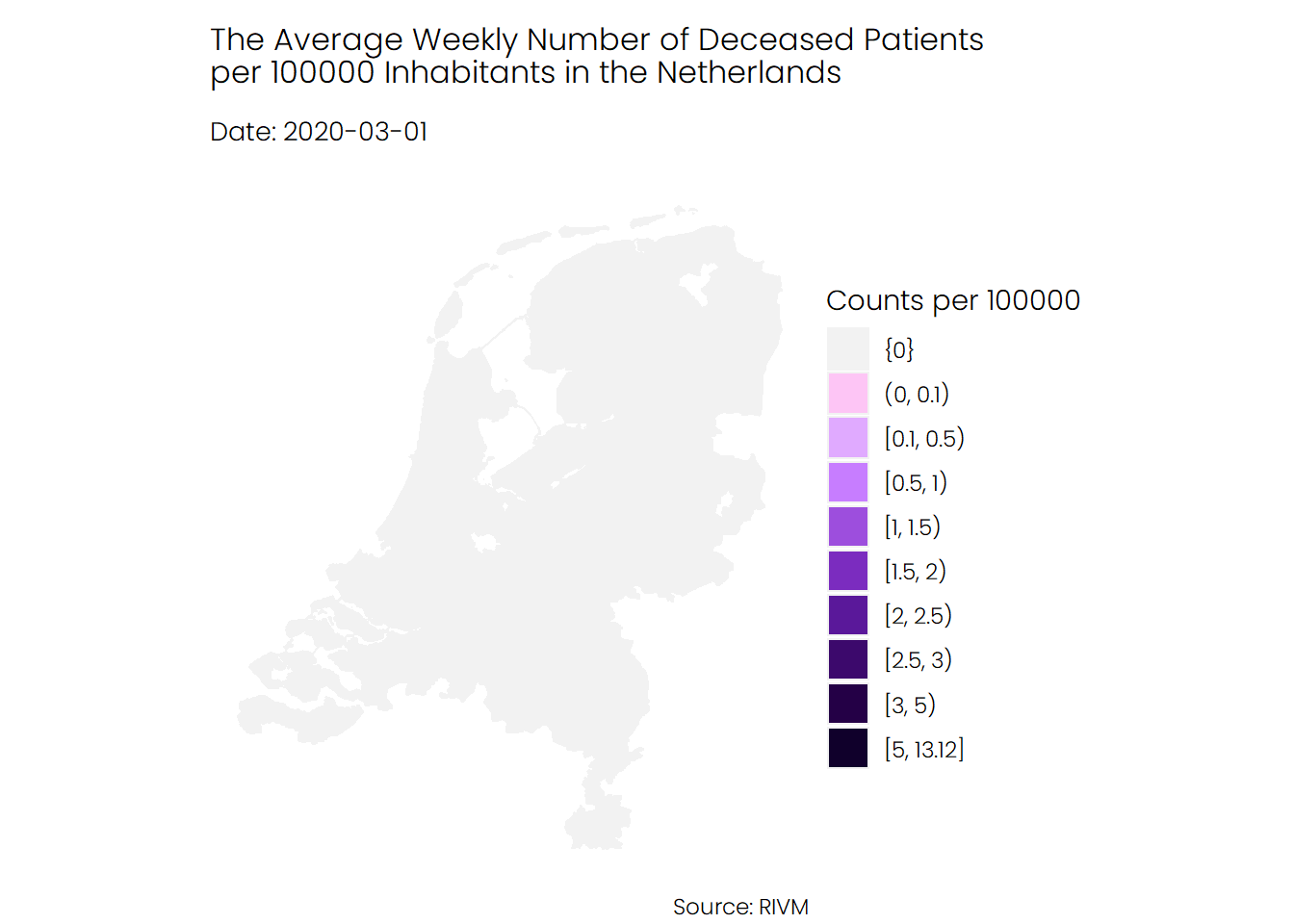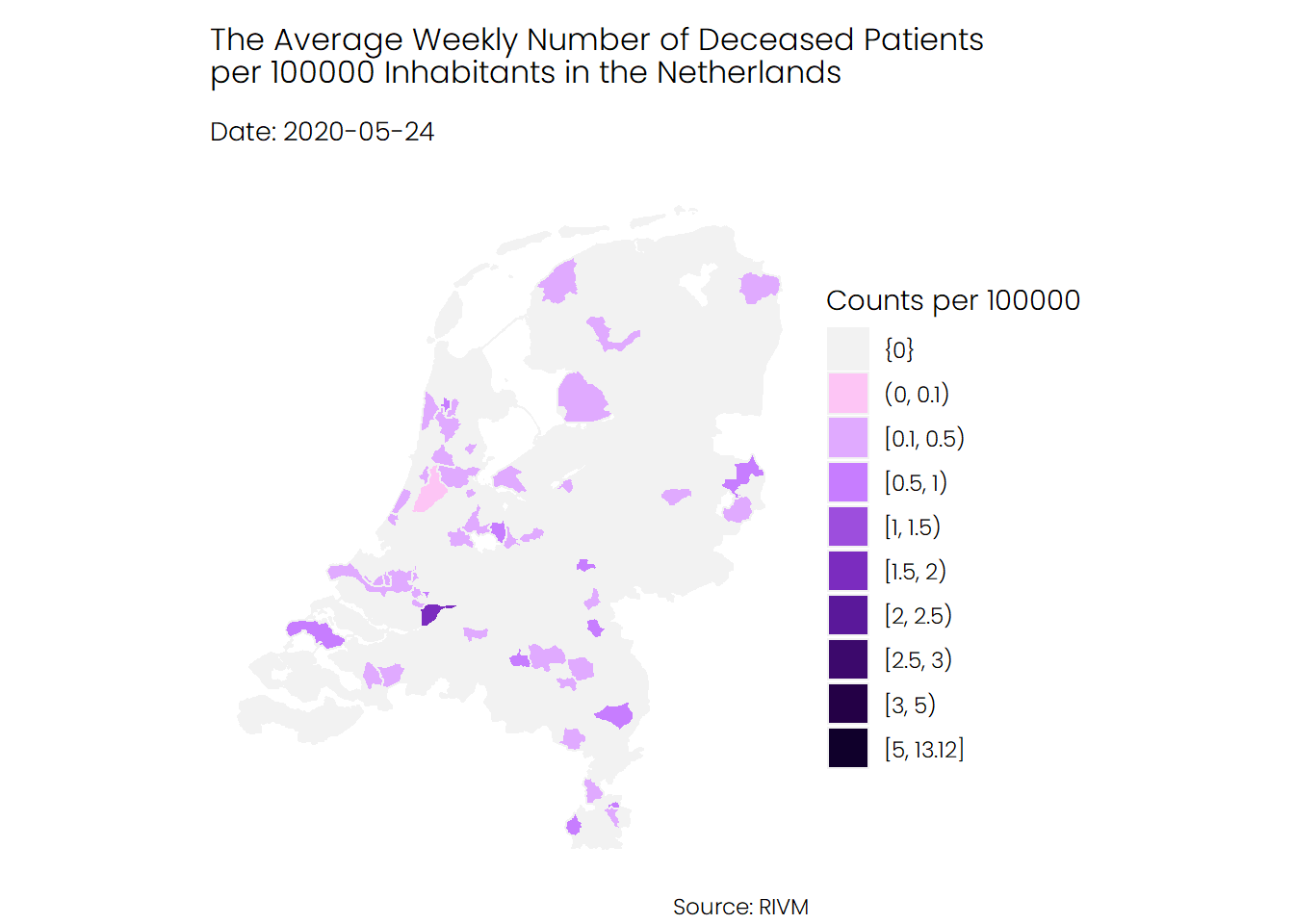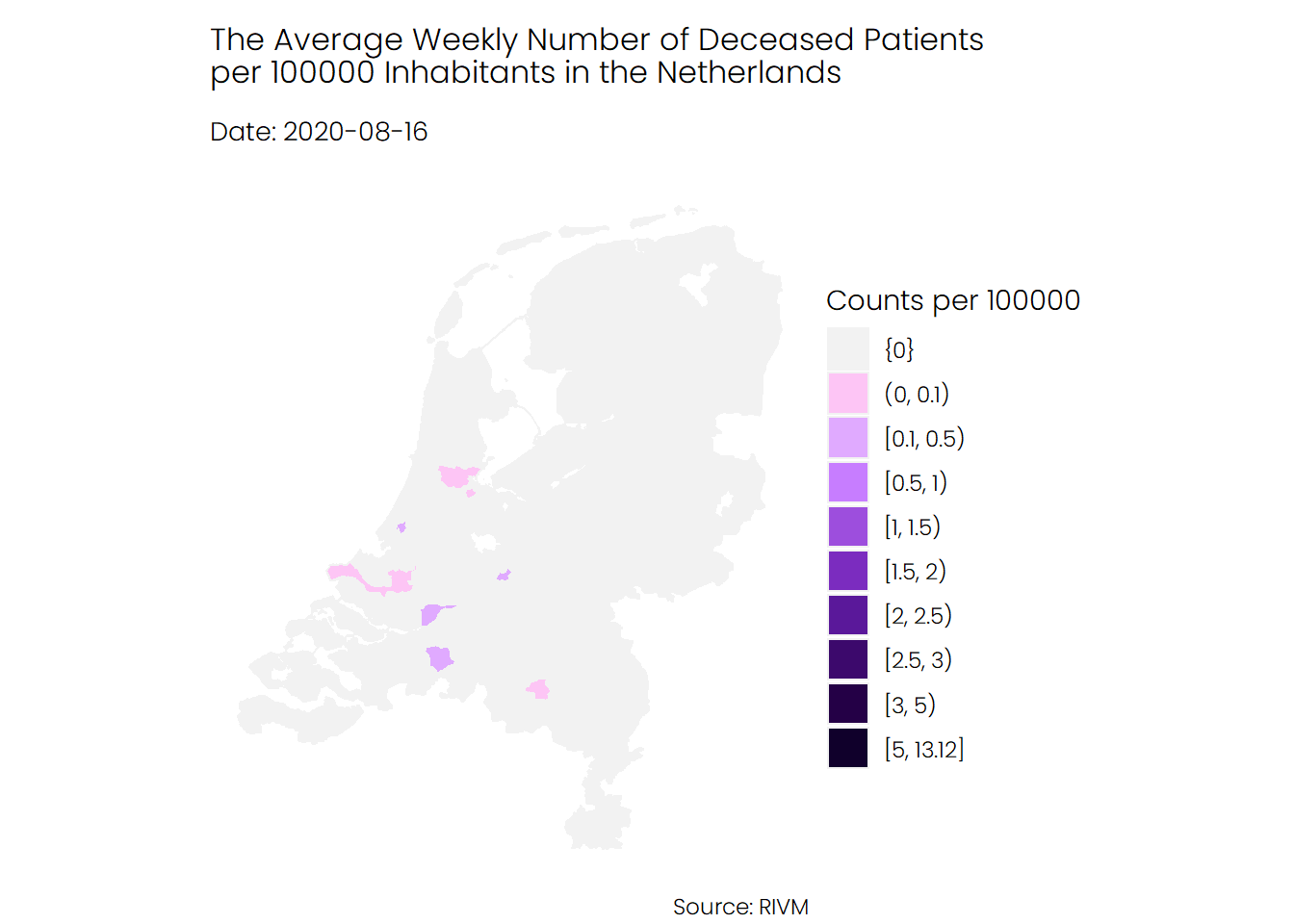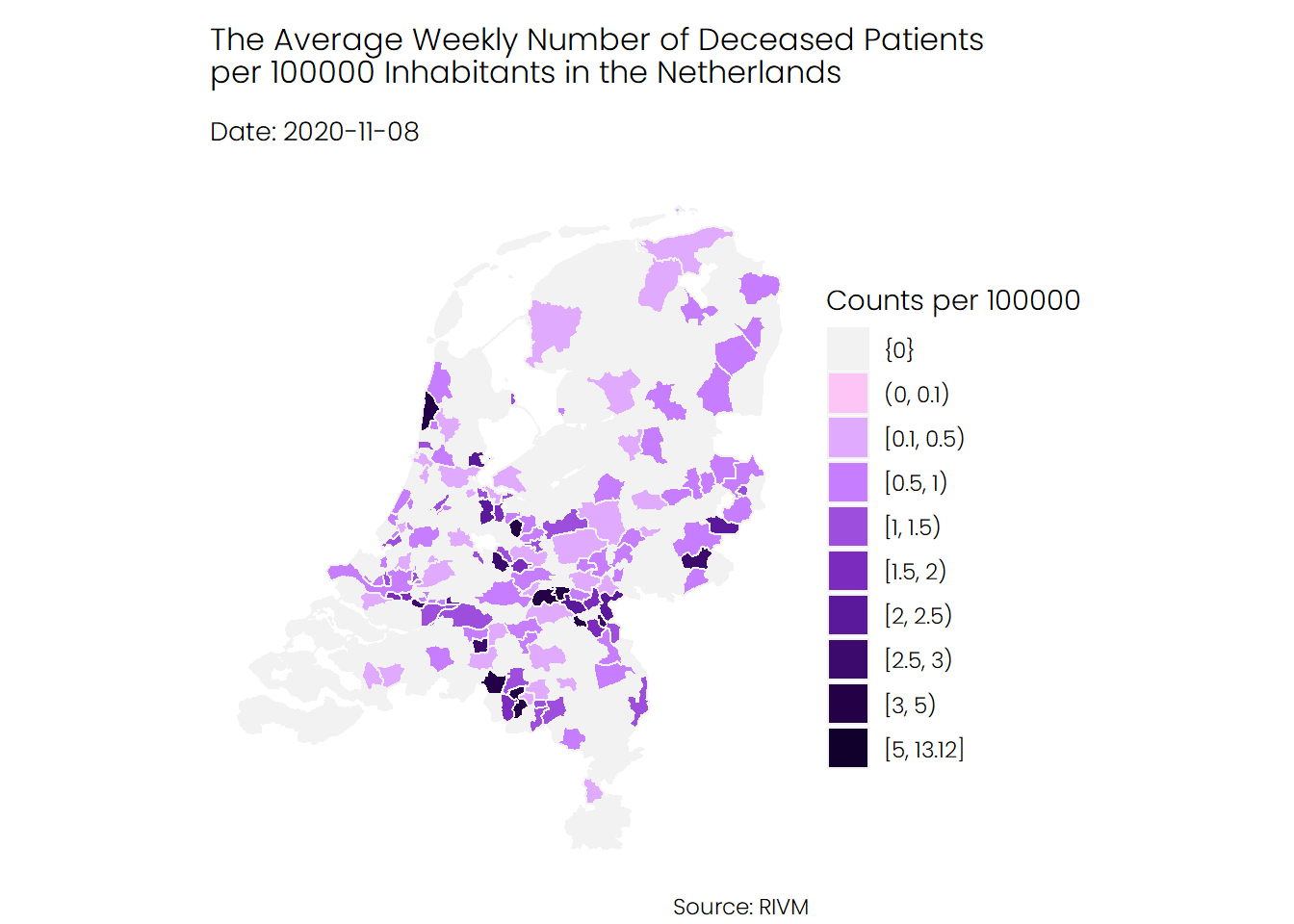
## Conclusion
These different animations show us two distinct trends. On the one hand, we can see the number of confirmed cases rose rapidly during the second wave to affect almost all regions. On the other hand, the number of hospitalizations and deaths during the second wave slightly decreased. This might suggest that the rise in the number of cases is mainly driven by an increase in the number of tests. Alternatively, the virus might have become less deadly and severe.


